
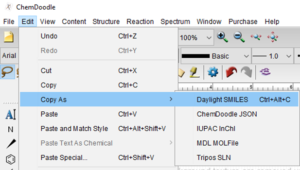

I like to calculate the theoretical amounts of all solids I’m supposed to weigh out, then go back and fill in the actual amounts as I weight them. Use formulas to make all of the calculations (e.g, for mmoles, just do =*/).Unit (narrow column-just big enough to fit “mg” or “µL”).Starting in cell B7, make columns for the following:.(Optional, but makes searching easier) Copy the structure as a SMILES string (Edit → Copy As → SMILES (⌥⌘C (Mac) or Ctrl + Alt + C (Windows)), and paste.Copy the text box near the structure containing the IUPAC name, formula, MW, and exact mass, and paste into the LabArchives rich text entry.Then, use View → Show Analysis Window → Paste to add formula, molecular weight, and exact mass to the document.In ChemDraw, select the product, and use the Structure → Convert Structure to Name command (⌥⌘N (Mac) or Ctrl + Alt + N (Windows)).Copy and paste from the Word document into a rich text entry in LabArchives.From there, copy and paste the structures and arrow into a Word document.Experimental write-up and characterization data.
While it may not offer all of the chemistry-specific bells and whistles that come with Perkin-Elmer’s offerings or the slick ease-of-use that comes with (more biology-focused) Benchling, LabArchives is a serviceable option for synthetic chemists with the right tricks.Ī good notebook entry for a reaction consists of three parts: Once you learn to use it, however, LabArchives isn’t half-bad. If you do organic synthesis research, you may be disappointed at first as you struggle with the silly chemical sketcher widget, complete lack of integration with ChemDraw, and weak stock spreadsheet widget. As a “jack-of-all-trades” electronic lab notebook, it tries to cater to every field of science. Let’s work together to make chemistry articles easier to find and use.LabArchives-love it or hate it, a growing number of academic institutions are adopting it ( reviewed here). Luckily, it really couldn’t be quicker or easier to improve the discoverability and reusability of your article by including machine-readable structure files or identifiers. The less time we have to spend re-drawing structures from pdfs, the more time we can devote to doing science. Press Ctrl+K, then select SMILES or InChI from the Copy As pop-upįrom the top menu, choose Edit > Copy As and select SMILES or InChI from the pop-upįinally, paste your SMILES or InChI into your document or spreadsheet. Right click, and choose Molecule > Copy As > SMILES or InChIįrom the top menu, choose Tools > Generate > SMILES Notation or InChI for Structure Avogadroįrom the top menu, choose Edit > Copy As > SMILES or InChIįrom the top menu, choose Edit > Copy As > Daylight SMILES or IUPAC InChI Start by selecting the structure you would like to copy as SMILES or InChI. The V3000 mol file has some extra features, but is not universally supported, so it is advised that you use V2000 mol format to ensure maximum interoperability.
#COPY AS IMAGE CHEMDOODLE SOFTWARE#
If there is more than one option, please be aware that V2000 mol format is more common and is supported by all cheminformatics software packages. Please note: There may be more than one molfile format listed in the dropdown. Select “MDL Molfile”, “MDL SDFile”, or “.mol” or “.sdf” in the dropdown. They generally follow the same steps:Ĭhoose File > Save As from the top menu OR press Ctrl+Shift+S. Save as MOL fileĪll major structure drawing packages can save structures as MOL files. Including these files or identifiers in your article or supplementary information helps make your article indexable and structure-searchable, and is a great way to make your article stand out. If you’re already drawing a structure for an article you are preparing to submit, it only takes a few seconds to generate machine-readable mol files or structure identifiers like SMILES or InChI.
Interested in making your article more discoverable and usable? As a reader, you have probably spent a lot of time re-drawing structures from an image in a PDF, or have struggled to find all relevant articles because your compound of interest is called by different names in different articles (IUPAC name, trivial name, registry number, drug development ID, generic name, brand name, revised trivial name etc etc etc…).


 0 kommentar(er)
0 kommentar(er)
